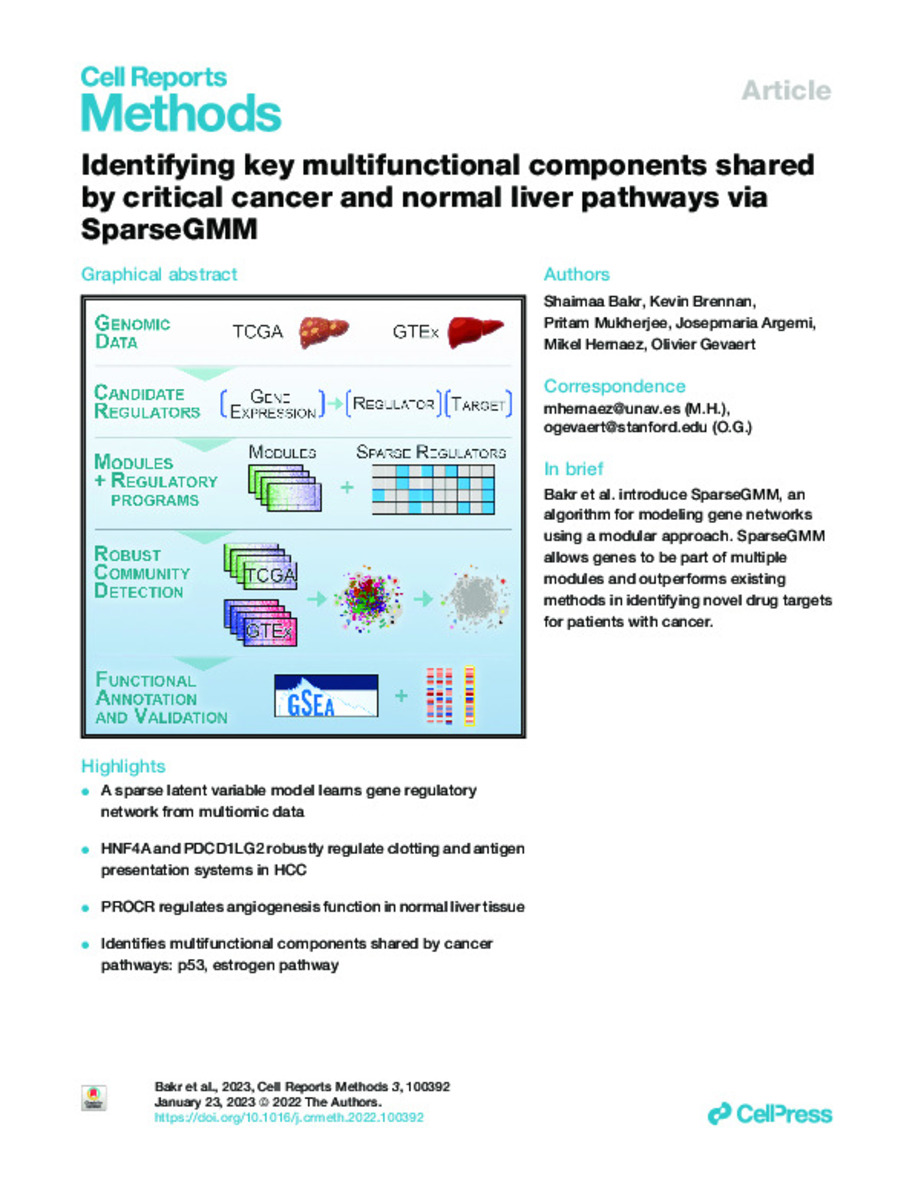
Identifying key multifunctional components shared by critical cancer and normal liver pathways via SparseGMM
Note:
This is an open access article under the CC BY-NC-ND license
Citation:
Bakr, S.; Brennan, K.; Mukherjee, P.; et al. "Identifying key multifunctional components shared by critical cancer and normal liver pathways via SparseGMM". Cell Reports Methods. 3 (1), 2023, 100392
Statistics and impact
0 citas en

0 citas en

Items in Dadun are protected by copyright, with all rights reserved, unless otherwise indicated.